Image credit Bristol University Department of Chemistry
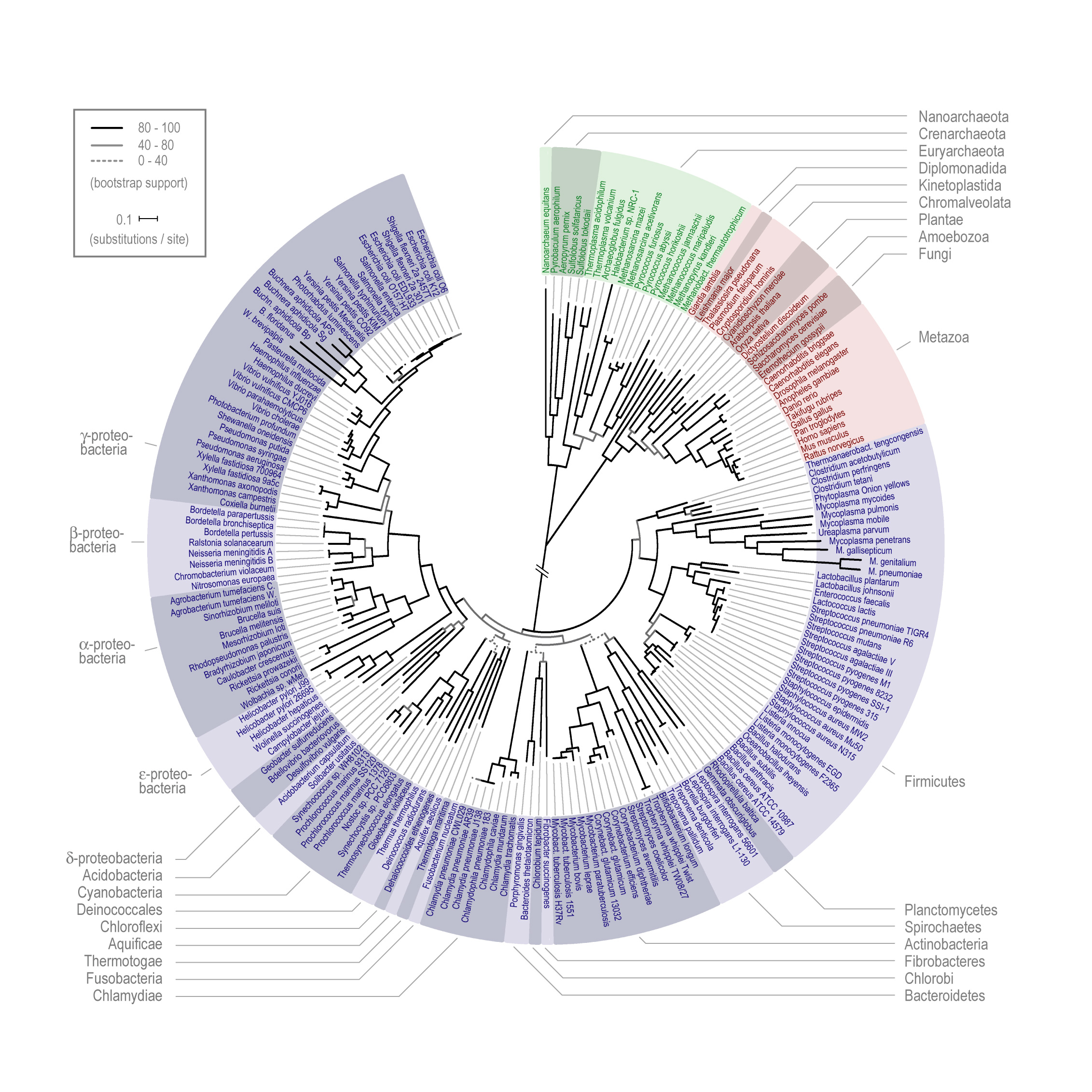
What would be really cool, and may already exist, which is why I'm posting this, is the same tool existed, except for gene ancestry. (And here is where I make obvious my ignorance of bioinformatics and the descent of genes both.) Genes can often be shown to originate by duplication events. I'm probably being naive about the extent to which the ancestry of genes results from similar events, or can be traced back to common ancestors shared with other genes in the same genome. But unless there was a lateral transfer event, genes all have to have common ancestry with other sequences in the organism, correct?
Ultimately we'd see another radial circle, except with genes radiating out from a common ancestor. The outer surface would be the human genome as it is now. You could place nested concentric circles representing previous phases of the genome (see mock-up below). Yes, I know the "surface" separating primates from non-primates (for example) doesn't represent any qualitative break, and the circle would be pretty lumpy since not all genes or gene families would branch and mutate at the same rates. (Genes I picked are illustrative only.)
Now, there are lots more genes than you could legibly read on a computer-screen-sized wheel, but this is also true for the species-descent wheel, which uses representative organisms. In my dream app, you could select certain gene families or genes with products that catalyzed certain classes or reactions or interacted with other gene products. The point of such a visualization is that you could more easily see when pathways started appearing or becoming more complex. Of course there will have been lots of genes lost in the interim, so we couldn't get a complete picture of what the genome was like at each point in the past. (Going back to the Grand-Daddy autocatalytic RNA would be cool, but unlikely.) If the gene-product features are sufficiently advanced you could even conceivably find "holes" in pathways at ancestral stages where the must have been gene products whose genes are no longer in the genome.
If there are any tools similar to this already out there, I would greatly appreciate a comment. This would be a valuable tool to investigate the evolution of pathways and systems. Especially neurons, of course.

No comments:
Post a Comment